Programmatically Retrieve Flat File MetaData by Python
A fair number of ETL tasks for Business intelligence developers are to load raw flat files. For large scale flat file loading, BIML provides a good solution to be able to generate massive packages programmatically. The following simple talk post showed how elegant the solution could be.
https://www.red-gate.com/simple-talk/sql/ssis/developing-metadata-design-patterns-biml/
However, this approach has a limitation that we will need to create the flat file format in advance in order for it to work. I have checked the BIML document if there is an easier way to create this format automatically, but I don’t have luck yet. Imagining you try to go through each flat file and insert the proper data type for each column, that will be painful.
To get accurate data type and length has a big impact on the database. It will save the space (int vs varchar) and make data accurately reflect what they really mean (varchar vs datetime).
I have seen different ways to handle it. For example, you can load the raw data in a staging database with varchar data type, then you will manually check the data and give the appropriate date type in the destination database. Some ETL tools will scan the first N rows of a flat file to determine the data type and length. The first method is labor intensive, the second method is automatic, but it only scans the first N row, when you load real data, you could have truncation or overflow error because there could be some records that are out of range after the first N row.
Do we have a better way to do it yet avoid the above pitfall?
The goal for this question is we need to have:
- Detect right data type automatically, no manual work.
- Give the adequate length if data type is text nature (this means we need to scan the full file)
- Automatically generate a column data type file to be used later (for e.g., BIML)
Please Note: if you are interested in usage of BIML, I have a blog talking about it at the following address. Link
Python has rich libraries and has been used in many fields to simplify the repetitive task. Let us see how Python can help us to reach the goal we defined.
The overall idea is as follows,
- import flat file to pandas dataframe
- Dataframe has dtypes property for each column. It will roughly tell you the data type, which is not always accurate.
- There are int8, int16… in python, but all start with int, same for float, object data type are a bit tricky. Because not all datetime will be following python datatime format, so many of them are classified as object data type. We will try a different approach to see if it is datetime.
- Need to implement multiple file scenario
- Need to output to a final file.
Let us get to how I implement this.
Import necessary library, as I commented the usage of each library
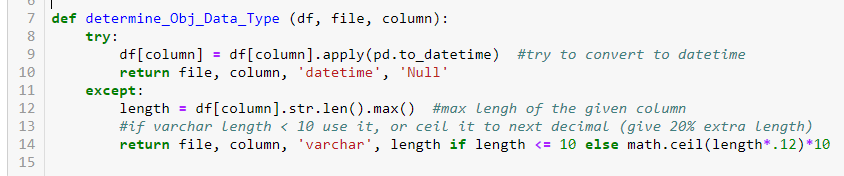
Pre-created a helper function to determine if an object data type column is a datatime or real string column. I have tried different approaches, but it is more reliable by trying to convert the given object column to datetime. If it went through, it was datetime datetype. If it did not, it was varchar by taking use of try except block.
Next is the main function that we are going to use to get the final result.
The function needs two parameters. One is a flat file source folder. The other is output flat file path, which will be your result. I will walk you though the key steps
Row 11: Use os library listdir function to get a list of files in this folder.
Row 12: only get file with appendix csv or txt
Row 13: use with statement to open the destination file in write mode. Please note: the directory must be there for use. But you don’t have to create the file there. The process will create the file.
Row 14-15: create writer object and write the column header to the file.
Row 16: loop through each file in the flat file collections
Row 18: read flat file into dataframe, please note: it use different format depending on csv /txt
Row 19: convert dataframe dtypes result to another dataframe and reset index because by default the results use column name as index.
Row 20: change column name of new data frame
Row 21-28 : iterate each row in the new data frame, if it is intXX ,write int, if it is floatXX, write float, if it is object, call the previous function to determine if it is datatime or real string.
Let us give a test drive.
I randomly downloaded some data from data.gov to do a test.
Three datasets are used, ranging from demographis, financial to police report. Largest row count is 22K. The widest table contains 70 columns. I think it might represent the majority of files we will come across on the daily basis.
I passed the directory and destination file to the function and calculated the time it needed to process the file. It takes about 5 sec to process a total 154 columns from 3 files. The performance is good for my requirements.
This is the result which shows in final_meta_data.
I did a brief check on the detection result with raw data. I think it did a good job.
The output can be easily convert to the format that BIML can use it to massively create SSIS package to load the data to relational database.
I hope you feel this function is helpful.
As always, I will include the download link here for Jupyter notebook and source files and output files.
Thanks and keep safe in 2021.
Wenlei